|
Protocol
for Quantitation of GFP and GFP Variants
1.
Introduction
The
green-fluorescent protein (GFP) from the jellyfish Aequorea
victoria is used as a fluorescent marker for gene expression
in a variety of organisms ranging from bacteria to higher
plants and animals.(1) The cloning of the gene for GFP
(2, 3) and its subsequent expression in heterologous
systems(1, 3, 4) has established GFP as a unique genetic
reporter system. It has become useful for monitoring
gene expression in vivo, in situ, and in real time.
When expressed in either eukaryotic or prokaryotic cells,
GFP gives forth a bright green fluorescence.
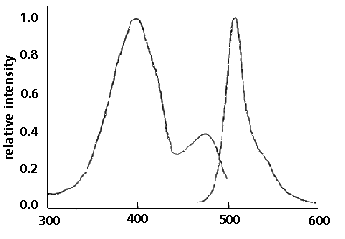
Figure 1. GFP emits green light (absorbance maximum
at 509 nm) when excited with blue or UV light (absorbance
maximum at 395nm). (1)
Unlike
other bioluminescent reporters, GFP fluoresces in the
absence of any other intrinsic or extrinsic proteins,
substrates, or cofactors. Fluorescence is stable, species-independent,
and can be monitored noninvasively in living cells and
whole animals, such as in the case of transparent organisms.(1)
Purified
GFP (from either native or recombinant sources) is a
27-kDa monomer consisting of 238 amino acids(2) . In
the bioluminescent jellyfish Aequorea victoria, light
is produced when energy is transferred from the Ca2+
activated photoprotein aequorin to GFP.(5, 6, 7). After
being formed, the protein appears to undergo an autocatalytic
reaction creating a fluorophore(8) from a backbone sequence
of amino acids, near residue number 66, which is a tyrosine.
A
putative GFP chromophore structure was described in
1979, and has since been modified(8, 5) (see Figure
2). This structure is unique amongst natural chromophores,
comprising a cyclic tripeptide sequence, serine-dehydrotyrosine-glycine,
which is covalently linked through the protein's peptide
backbone. The mechanism of chromophore formation is
unknown, but oxidation of tyrosine to dehydrotyrosine
is probably an essential step that occurs gradually
after the formation of the protein.
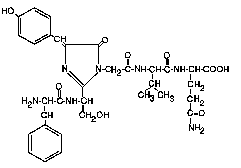
Figure 2. The Green-Fluorescent protein chromophore.(8)
2.
Materials Required
- TD-700
Fluorometer with standard PMT (P/N 7000-009)
- 13
mm round test tube adaptor (P/N 7000-981)
- 13
mm x 100 mm borosilicate glass test tubes (P/N 10-031)
- Tris-EDTA
buffer pH 8.00 (10 mM tris/10 mM EDTA/0.02% Sodium
Azide (w/v), adjusted to pH 8.00)
- 20µg
solution of recombinant Aequorea Green-Fluorescent
Protein from CLONTECH Laboratories, Inc. Telephone
(415)424-8222 (catalogue # 8360-1)
- GFP
(395 nm ex, 509 nm em) and GFPuv (360 - 400
nm ex, 509 nm em): Near UV Mercury Vapor lamp (P/N
10-049), 390 nm excitation filter (P/N 034-0390),
510 nm - 700 nm emission filter (P/N 10-109R-C)
- EGFP
(490 nm ex, 505 nm em) and EYFP (488 nm ex,
527 nm em): Blue Mercury Vapor lamp (P/N 10-089),
486 nm excitation filter (P/N 034-0486), 510 nm -
700 nm emission filter (P/N 10-109R-C)
- EBFP
(380 nm ex, 440 nm em): Near UV Mercury Vapor lamp
(P/N 10-049), 390 nm excitation filter (P/N 034-0390),
410 nm - 600 nm emission filter (P/N 10-110R-C)
3.
Standard preparation
3.1
The first step in quantitating GFP in solution is
to prepare a standard solution of highly purified
recombinant GFP.
3.2
A 20µg aliquot of highly purified recombinant
GFP is diluted into a final volume of 4 ml with 10
mM Tris-EDTA buffer (TE) containing azide at pH 8.00
to produce a 0.005 mg/ml standard solution (5µg/ml).
3.3
This is placed into a labeled 13 x 100 mm borosilicate
test tube and sealed with parafilm. Solutions of this
concentration are routinely used in the GFP field
and are quite suitable to calibrate fluorometers such
as the TD-700.
4.
Fluorometer Calibration
4.1
Considerations
4.1.1
Upon receiving and unpacking your TD-700 you must
prepare it for GFP quantitation. Identify the operating
manual and keep it within reach throughout this
operation.
4.1.2
Follow the procedure outlined in your operation
manual to install the near UV mercury vapor lamp
which is shipped outside the unit.
4.1.3
Install your GFP filters into the filter cylinder
and place it into the fluorometer (remember to wipe
any fingerprints off of any filter or cuvette after
you handle it). It will help to use the diagrams
in your TD-700 manual as a guide. The ports for
each set of filters are labeled EX for excitation
and EM for emission and each pair of ports is labeled
A through D. Choosing one pair of ports, carefully
insert your filters. The 390 nm filter has a reflective
face which should be installed so that it will face
out towards the lamp. Each filter is held in place
by a circular rubber grommet, or o-ring.
4.1.4
At the ends of the filter cylinder are labeled marks
corresponding to the pair of filter ports you have
chosen. Insert the filter cylinder into the fluorometer
while aligning this mark with the silver alignment
mark found on the inside rim of the fluorometer's
sample chamber.
4.2
Calibration
4.2.1
Close the TD-700 lid and turn the unit on. The
on/off switch is diagrammed in your manual. It will
count down 600 seconds to warm up.
4.2.2
After the instrument warms up, insert the cuvette
holder into the sample chamber. Note that the top
of the cuvette holder has an arrow shape molded
to the top part of the holder. Orient this arrow
pointing toward the silver alignment mark on the
inside rim of the sample chamber.
4.23
You will be performing a simple mode calibration
(refer to your manual if need be). Press [enter]
from the keypad. Enter [1] on the keypad to enter
"setup", then enter [1] to enter "mode".
Using the [arrow] key to choose the mode, select
"simple", then enter [ESC] twice to return
to the setup/calibration screen. Enter [2], then
press [enter]. The TD-700 will prompt you to insert
a typical sample. Place the test tube with your
standard GFP solution inside the chamber, close
the lid, and press [enter]. The TD-700 will now
automatically set the standard to read 500 out of
a total reading of 1,000 (the standard is set to
50% of maximum). When the screen indicates that
the sample is 500, press [enter]. Your TD-700 is
now calibrated.
5.
Quantitating GFP
5.1
Place the sample to be assayed for GFP into a 13 x
100 mm borosilicate test tube. Make sure that the
volume in the tube is over 3 ml. Anything under this
volume will not clear the slits on the cuvette holder
and the meniscus of the sample will interfere with
excitation and emission.
5.2
Place the tube into the chamber and close the lid.
The unit will measure the sample instantly.
5.3
Wait approximately 20 seconds for the signal to stabilize
before recording it. If the output is 1,000 or over,
the unit will read "OVER" indicating that
the sample is too concentrated to read at the current
sensitivity level. If this occurs, it is necessary
to dilute your sample and perform another reading.
Note:
The signals on fluorometers tend to drift over time,
always insert your standard and record its value,
(which is never far from 500) before taking any reading.
6.
Example Quantitation
You
have a strain of yeast expressing GFP under the sole
control of a promoter used to normally control an
enzyme that detoxifies a carcinogen. You grow a 10
liter culture of this strain to a predefined cell
density, expose it for a period of time to a known
concentration of this carcinogen, collect the cells,
and prepare a cell extract. This extract is at a final
volume of 1 liter. You prepare a 1 to 4 dilution of
this extract by diluting 1 ml of the extract to a
final volume of 4 ml with 20 mM Tris at pH 8.00. You
mix this and derive a reading of 663 on your TD-700.
Your standard was reading 510 at the time of the reading.
You perform the following calculation to determine
the amount of GFP induced in that exposure period
for that amount of cells.
[(sample
reading x dilution)/standard reading]
x 0.005 mg/ml x volume of extract
in
this case
[(663 x 4)/510] x 0.005 mg/ml x 1000 ml
=
26 mg of GFP produced during the period of exposure
in 10 liters of yeast culture.
REFERENCES
- Chalfie,
M., Tu, Y., Euskirchen, G., Ward, W.W., and Prasher,
D.C., © 1994, Green Fluorescent Protein as a Marker
for Gene Expression. Science. 263:802-805.
- Prasher,
D.C., Eckenrode, V.K., Ward, W.W., Prendergast, F.G.,
and Cormier, M.J., © 1992, Primary structure of the
Aequorea victoria green-fluorescent protein. Gene.
111:229-233.
- Inoue,
S., and Tsuji, F.I. © 1994, Aequorea green-fluorescent
protein: Expression of the gene and fluorescence characteristics
of the recombinant protein. FEBS Letters. 341:277-280.
- Wang,
S., and Hazelrigg, T. © 1994, Implications for bed
mRNA localization from spatial distribution of exu
protein in Drosophila oogenesis. Nature. 369:400-403.
- Shimomura,
O., Johnson, F.H., and Saiga, Y., © 1962, Extraction,
purification and properties of aequorin, a bioluminescent
protein from the luminous hydromedusan, Aequorea.
J. Cell. and Comp. Physiol. 62:1-8.
- Morin,
J.G., and Hastings, J.W., © 1971, Energy Transfer
in a Bioluminescent System. J. Cell Physiol. 77:313-317.
- Ward,
W.W., Cody, C.W., Hart, R.C., and Cormier, M.J., ©
1980, Spectrophotometric identity of the energy-transfer
chromophores in Renilla and Aequorea green-fluorescent
proteins. Photochem. Photobiol. 31:611-615.
- Cody,
C.W., Prasher, D.C., Westler, W.M., Prendergast, F.G.,
and Ward, W.W., © 1993, Chemical Structure of the
Hexapeptide Chromophore of the Aequorea Green-Fluorescent
Protein. Biochemistry. 32(5):1212-1218.
About the Author
This
GFP application note was written by Daniel G. Gonzalez
M.S., who is currently a Biochemistry Ph.D candidate
working in the laboratory of William W. Ward at Rutgers
University (New Brunswick, N.J.). His interests include
the physical characterization of chromophore formation
in a variety of Green-Fluorescent proteins from various
organisms. William W. Ward is one of the pioneers
in GFP research and is still very active in the field.
Both he and Daniel currently use a wide assortment
of techniques in fluorescence analysis, protein purification
and molecular biology to study and characterize Green-Fluorescent
proteins.
The
Ward lab currently prepares and coordinates a series
of short courses in biotechnology that features Aequorea
GFP as a model protein for purification and molecular
manipulation. For information on these courses, contact
Daniel at: Phone: (908) 932-9071, ext. 212, e-mail:
meton@rci.rutgers.edu,
Mailing address: Daniel Gonzalez, Rutgers University,
Cook College, Biochemistry and Microbiology, New Brunswick,
NJ 08903-0231.
Daniel
wishes to thank Turner BioSystems for making this
applications note possible and for providing the GFP
community with a useful analysis tool in their TD-700
fluorometer.
|

