|
Protocol
for DNA Quantitation
1.
INTRODUCTION
The
Turner BioSystems TD-20/20 Luminometer, in combination
with Promega’s new DNA Quantitation System, provides
a precise and sensitive method for the detection of
linear double stranded DNA (dsDNA) including PCR fragments.
Plasmid and chromosomal DNA can be quantitated following
linearization. The system is unaffected by many of
the contaminants that affect other systems. The measurement
is based upon a series of coupled enzymatic reactions
that produce a light signal proportional to the amount
of linear DNA in a sample.
The
DNA Quantitation System is more precise than spectrophotometric
or agarose gel analyses and is able to detect picogram
amounts of DNA. The assay should be used in the linear
range of 20pg to 1ng of total DNA added per reaction
and can quantitate linear dsDNA species up to 6,000bp
in size. Estimation of DNA amount is dependent upon
mass and not the number of DNA ends or nicks in the
DNA for fragments < 6,000bp.
Since
the DNA Quantitation System depends upon the production
of ATP by enzymatic reactions, you must take extra
care to insure that the samples do not contain dNTPs
or NTPs, which can be used to form ATP in the reactions.
Unless a DNA sample is contaminant-free, a control
not containing T4 DNA Polymerase should be performed.
This control reaction will gauge the contribution
of signal from materials other than dsDNA. Furthermore,
the sample should not contain any ATPase activity.
2.
MATERIALS REQUIRED
- TD-20/20
Luminometer (P/N 2020-000)
- 8mm
Test Tube Adapter (P/N 2020-314)
- 8mm
x 50mm test tubes (P/N 20-015)
- Microfuge
tubes (Sterile)
- Micropipettes
(Aerosol Resistant Tips are Recommended)
- Sterile
10mM Tris-HCl (pH 7.3-7.5)
- Sterile
TE buffer (pH 7.3-7.5)
- Sterile
deionized water
- DNA
Quantitation System containing:
2
x 1ml, DNA Quantitation Buffer Solution containing
ADP
2
x 500u, T4 DNA Polymerase (5-10 u/µl)
50
µl, Sodium Pyrophohsphate (40mM)
15µl,
NDPK Enzyme Solution
200µl,
DNA Quantitation DNA Standard (5ng/µl ± 5%)
1
vial, ENLITEN® Luciferase/Luciferin reagent
1
vial, ENLITEN® Reconstitution Buffer
(12ml)
3.
EXPERIMENT PROTOCOL
3.1
ENLITEN® Luciferase/Luciferin reagent
preparation: Use as supplied by Promega Corporation.
Store at -20° C. Lightly tap the vial of ENLITEN®
L/L Reagent before opening to gather all of the dried
material to the bottom of the vial. Transfer 12ml
of the ENLITEN® L/L Reconstitution Buffer
into the vial of ENLITEN® L/L Reagent.
Replace the stopper carefully, and gently invert the
vial several times to dissolve the contents. Do not
shake the dissolved ENLITEN® L/L Reagent.
Equilibrate the reconstituted ENLITEN®
L/L Reagent to room temperature for a minimum of 30
minutes and a maximum of 8 hours. Unused reconstituted
reagent should be dispensed into sterile microcentrifuge
tubes and frozen at -20° C
3.2
Reaction Master Mix Preparation: Thaw the enzyme and
buffer solution and keep on ice. Mix the reagents
well by gentle inversion or vortexing.
Note:
Prepare the reaction master mix just prior to
use. Determine the number of reactions to be performed
for samples and that to prepare a standard curve.
All samples and controls should be performed in duplicate
or triplicate.
Prepare
a master mix using Table 1 as a guide. Add the components
in the order listed to a fresh microcentrifuge tube,
mix gently and keep the solution on ice.
Number
of Reactions (n)
|
Component
|
1–9
|
³
10
|
Experiment
|
|
DNA
Quantitation
Buffer Solution
|
n
× 15.5µl
|
n
× 15.5µl
|
_
× __µl = ___µl
|
|
Sodium
Pyrophosphate
|
n
× 0.5µl
|
n
× 0.5µl
|
_
× __µl = ___µl
|
|
NDPK
Enzyme Solution
|
n
× 1.0µl
|
n
× 0.1µl
|
_
× __µl = ___µl
|
|
Water,
sterile deionized
|
—
|
n
× 0.9µl
|
_
× __µl = ___µl
|
|
T4
DNA Polymerase
|
n
× 1.0µl
|
n
× 1.0µl
|
_
× __µl = ___µ l
|
|
Total
Volume
|
n
× 18.0µl
|
n
× 18.0µl
|
_
× __µl = ___µl
|
Table
1. Volume of Master Mix Components Required by Number
of Reactions.
4.
INSTRUMENT SET-UP
4.1
Turn on instrument and allow to warm up for at
least 5 minutes (600 seconds).
4.2
Adjust settings so that:
Delay
period = 3sec.
Integrate period = 15sec.
Replicates = 1
5.
SAMPLE ANALYSIS
5.1
Prepare the reconstituted ENLITEN® L/L
Reagent. Allow to equilibrate to room temperature.
5.2
Prepare samples for the standard curve by making dilutions
of the provided DNA Quantitation DNA Standard. The
DNA standard should be mixed well. Dilutions can be
made using 10mM Tris-HCl (pH 7.3–7.5) or TE buffer
(10mM Tris, 1mM EDTA [pH 7.3–7.5]). The prepared
samples should span the region of interest in the
linear range of the curve.
5.3
If needed, dilute the DNA samples to be tested to
10–500pg/µl using 10mM Tris-HCl (pH 7.3–7.5)
or TE buffer.
5.4
Prepare the reaction master mix.
5.5
Aliquot 18µl of master mix into each prelabeled microcentrifuge
tube.
5.6
Add 2µl of DNA solution (from duplicate or triplicate
reactions) to each of the tubes and vortex gently.
Include a negative control containing 2µl of the buffer
used to dilute the standard.
5.7
Incubate at 37°C (in a heating block or water bath)
for 8–10 minutes. Do not incubate longer.
5.8
Place samples on ice and analyze as soon as possible.
5.9
Remove 15µl from one reaction mix and place into
100µl of reconstituted ENLITEN® L/L Reagent.
Mix gently.
5.10
Immediately read the sample by pressing "GO"
on the TD-20/20. Repeat Steps 9 and 10 until all samples
are read.
6.
DNA CONCENTRATION CALCULATION
6.1
Average duplicate or triplicate sample values.
6.2
Subtract the averaged value for the "no DNA"
blank reaction from that obtained for the other samples.
This yields net light units (LU).
Net
average LU (y) = Average LU sample – Average
LU No DNA Control
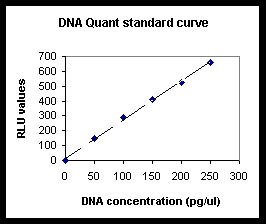
6.3
Generate the standard curve by plotting DNA concentration
vs. net light units and drawing a best-fit line (of
the form y = mx, or y = mx + b).
6.4
Determine the concentration of the unknown samples
by comparing the net light units seen for these samples
against those seen for the standard curve.
7.
RECOMMENDED READING
"DNA
Quantitation System," Technical Bulletin 278
(available from Promega
Corporation).
| 
